3. Forecasting with Time Series Models
Source:vignettes/03-times-series-models.Rmd
03-times-series-models.RmdSetup
Set parameters
state_name <- "Maryland"
geo_ids <- "md"
forecast_date <- as.Date("2024-12-01")
forecast_disease <- "influenza"
target <- "wk inc flu hosp"
forecast_horizon_wks <- 0:3Load packages
knitr::opts_chunk$set(echo = TRUE)
library(AMPHForecastSuite)
library(tidyverse)
library(forecast)
library(jsonlite)
library(epidatr)
library(epipredict)
library(epiprocess)
library(ggplot2)
library(dplyr)Load target (observed) data
We already pulled and saved the data we need in
Collect Empirical Data. See
vignettes/collect_empirical_data.Rmd for details.
# Load data saved from a specific forecast date:
target_data_path <- file.path("target-data", paste0("target-hospital-admissions-", forecast_date, ".csv"))
target_data <- readr::read_csv(file = target_data_path) %>%
mutate(location = as.character(location)) %>%
dplyr::filter(!is.na(observation)) %>% # keep rows with observed values
dplyr::arrange(dplyr::across(dplyr::any_of(c("target_end_date","date"))))
# get location id to add to forecast output
location_dat <- target_data %>%
dplyr::select(location, abbreviation, location_name) %>%
distinct()
location <- as.character(location_dat$location)Define Reference date
The reference_date is the Saturday for the week of the
forecast date. This is the date that will be used in the forecast
submission file.
reference_date <- get_reference_date(forecast_date)## Reference date: 2024-12-07## Forecast date: 2024-12-01Models
SARIMA (forecast package)
Here we use the auto.arima function from the
forecast package to fit a seasonal ARIMA model to the data
(seasonal = T). Use (seasonal = F) to remove seasonality.
Run the model
fc_sarima <- forecast::auto.arima(y = target_data$observation,
seasonal = T,
lambda = "auto") %>%
forecast::forecast(h = length(forecast_horizon_wks),
level = c(.1, .2, .3, .4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.95, 0.98))## Warning in guerrero(x, lower, upper): Guerrero's method for selecting a Box-Cox
## parameter (lambda) is given for strictly positive data.Plot SARIMA forecast results
# Build data for plotting
h <- length(forecast_horizon_wks)
# dates for history + horizon (uses target_end_date if present; else index)
hist_dates <- if ("target_end_date" %in% names(target_data)) as.Date(target_data$target_end_date) else seq_len(nrow(target_data))
fc_dates <- if (is.Date(hist_dates)) max(hist_dates) + 7L*seq_len(h) else max(hist_dates) + seq_len(h)
df_hist <- data.frame(date = hist_dates, value = target_data$observation)
pick <- function(mat, lvl){
nm <- paste0(lvl, "%")
if (!is.null(colnames(mat)) && nm %in% colnames(mat)) mat[, nm] else mat[, ncol(mat)]
}
df_fc <- data.frame(
date = fc_dates,
mean = as.numeric(fc_sarima$mean),
l80 = as.numeric(pick(fc_sarima$lower, 80)),
u80 = as.numeric(pick(fc_sarima$upper, 80)),
l95 = as.numeric(pick(fc_sarima$lower, 95)),
u95 = as.numeric(pick(fc_sarima$upper, 95))
)
# keep only the last 10 observed points to view most recent data
df_hist_last <- df_hist %>% arrange(date) %>% slice_tail(n = 10)
# x-range: last observed window through end of forecast
x_start <- min(df_hist_last$date)
x_end <- max(df_fc$date)
ggplot() +
geom_point(data = df_hist_last, aes(date, value)) +
geom_ribbon(data = df_fc, aes(date, ymin = l95, ymax = u95), alpha = 0.15) +
geom_ribbon(data = df_fc, aes(date, ymin = l80, ymax = u80), alpha = 0.25) +
geom_line(data = df_fc, aes(date, mean)) +
coord_cartesian(xlim = c(x_start, x_end)) +
labs(title = "SARIMA model fit and forecast",
x = "Date", y = "Hospital admissions") +
theme_minimal(base_size = 12)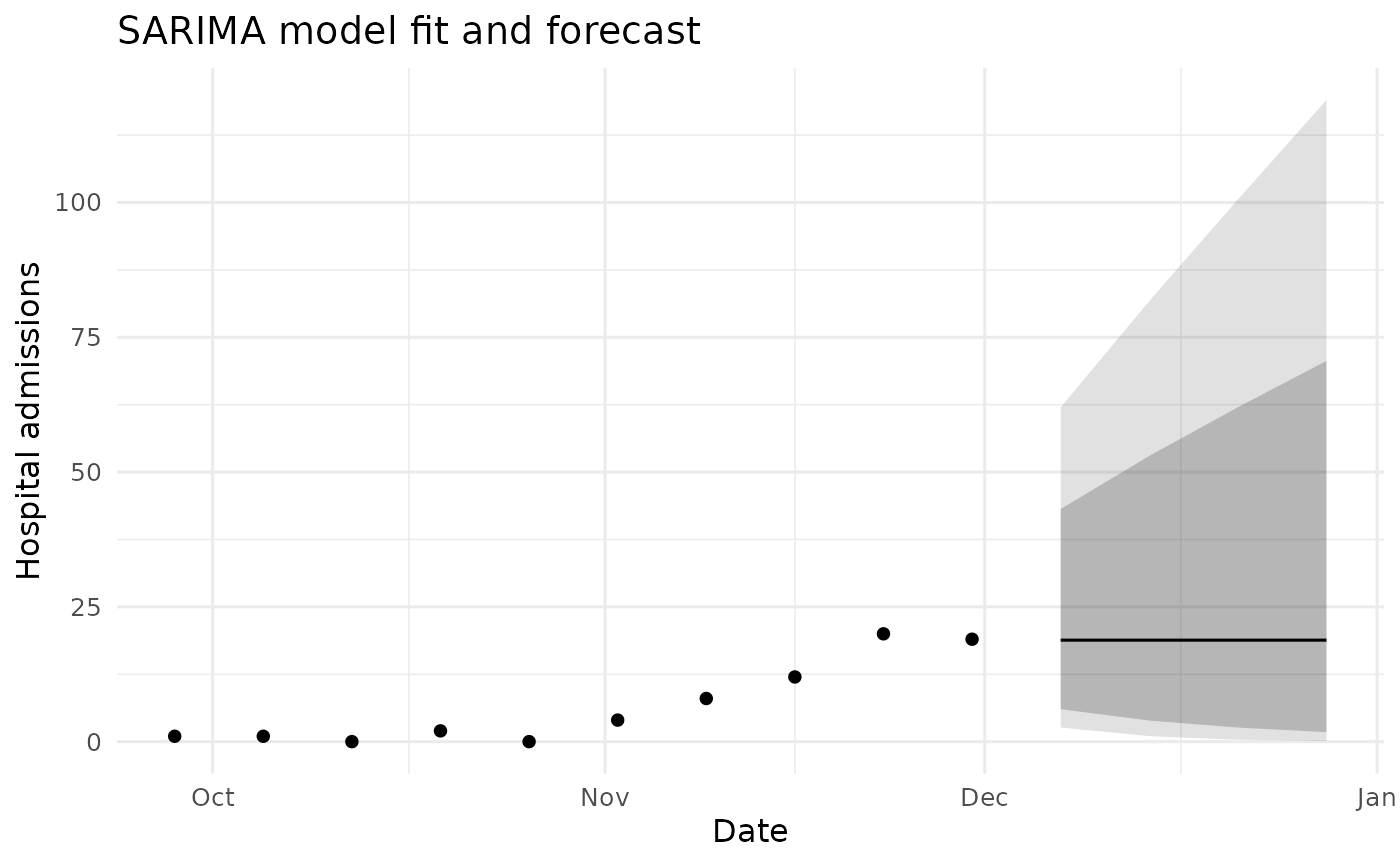
SARIMA model fit and forecast
Check that the model output is in the correct format.
We use the hubValidations package to check against the
tasks.json file we create in
Getting Started.
# Future functionality. Not yet developed.Save in hubVerse Format
# Transform to hubVerse format
data_fc_sarima <- trans_forecastpkg_hv(fc_output = fc_sarima,
model_name = model_name,
target = target,
reference_date = reference_date,
horizon_time_steps = forecast_horizon_wks,
geo_ids = location)
## Save data file
#' -- this will have validation build in for fluid workflow eventually.
save_model_output(model_name = model_name,
fc_output = data_fc_sarima,
reference_date)## Model output saved to model-output/AMPH-sarima/2024-12-07-AMPH-sarima.csvNeural network model (forecast package)
Here we use the nnetar function from the
forecast package to fit a neural network model to the
data.
Run the model
fc_nnet <- forecast::nnetar(target_data$observation,
lambda = "auto") %>%
forecast(PI = TRUE,
h = length(forecast_horizon_wks),
level = c(.1, .2, .3, .4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.95, 0.98))## Warning in guerrero(x, lower, upper): Guerrero's method for selecting a Box-Cox
## parameter (lambda) is given for strictly positive data.Plot Neural Network forecast results
df_fc <- data.frame(
date = fc_dates,
mean = as.numeric(fc_nnet$mean),
l80 = as.numeric(pick(fc_nnet$lower, 80)),
u80 = as.numeric(pick(fc_nnet$upper, 80)),
l95 = as.numeric(pick(fc_nnet$lower, 95)),
u95 = as.numeric(pick(fc_nnet$upper, 95))
)
ggplot() +
geom_point(data = df_hist_last, aes(date, value)) +
geom_ribbon(data = df_fc, aes(date, ymin = l95, ymax = u95), alpha = 0.15) +
geom_ribbon(data = df_fc, aes(date, ymin = l80, ymax = u80), alpha = 0.25) +
geom_line(data = df_fc, aes(date, mean)) +
coord_cartesian(xlim = c(x_start, x_end)) +
labs(title = "Neural network model fit and forecast",
x = "Date", y = "Hospital admissions") +
theme_minimal(base_size = 12)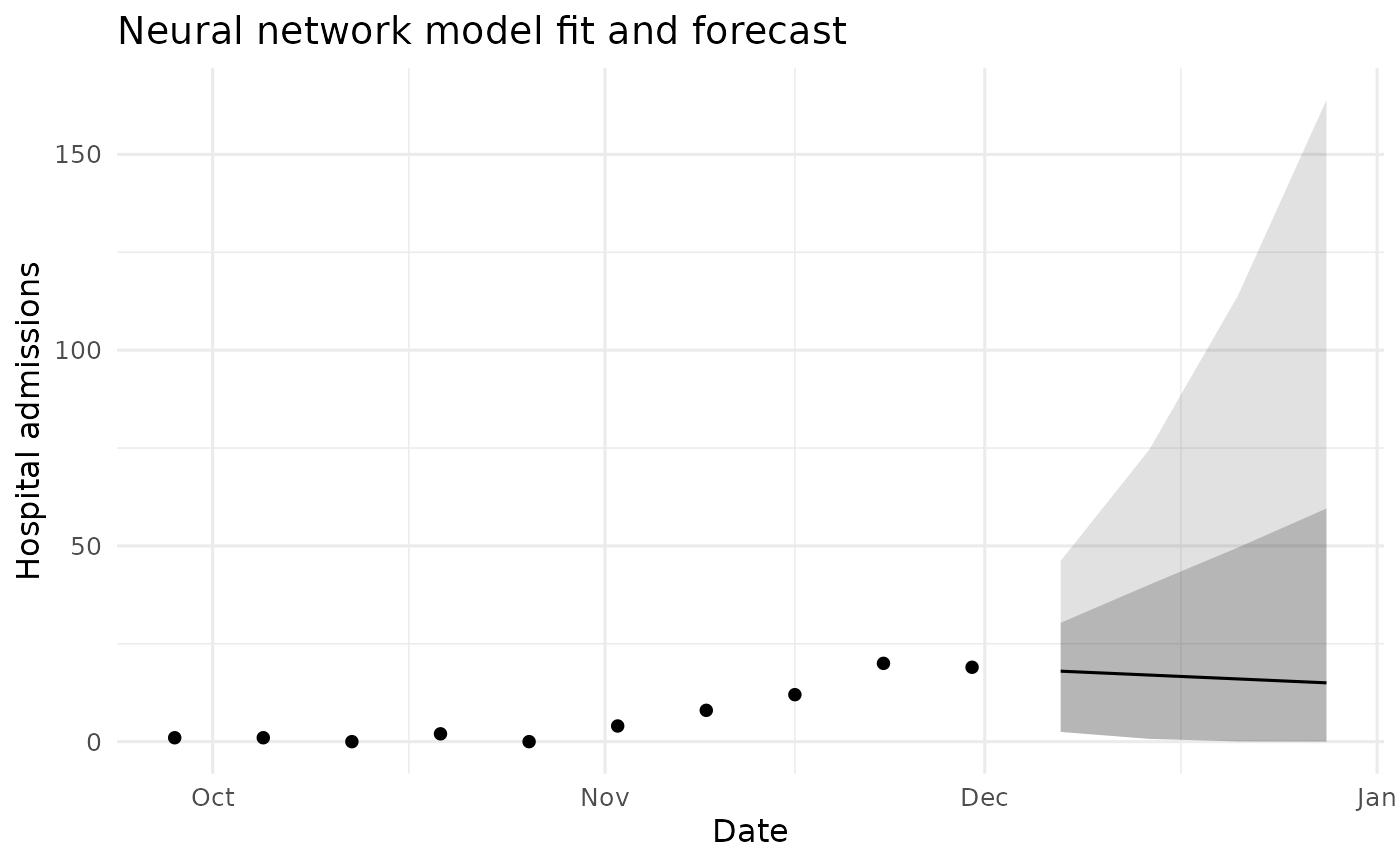
Neural network model fit and forecast
Save in hubVerse Format
# Transform to hubVerse format
data_fc_nnet <- trans_forecastpkg_hv(fc_output = fc_nnet,
model_name = model_name,
target = target,
reference_date = reference_date,
horizon_time_steps = forecast_horizon_wks,
geo_ids = location)
## Save data file
#' -- this will have validation build in for fluid workflow eventually.
save_model_output(model_name = model_name,
fc_output = data_fc_nnet,
reference_date)## Model output saved to model-output/AMPH-neuralnetwork/2024-12-07-AMPH-neuralnetwork.csvAutoregressive Forecaster (epipredict package)
Here we use the epipredict package to fit an
autoregressive model to the data. We specify lags of 0, 7, and 14 days
(i.e., current week, previous week, and two weeks ago) and forecast
horizons of 7, 14, 21, and 28 days ahead (i.e., 1 to 4 weeks ahead).
Run the model
# Set up data for epipredict
target_data_arx <- target_data %>%
rename(geo_value = location,
time_value = target_end_date,
value = observation) %>%
tsibble::as_tsibble(index = time_value, key = c(geo_value)) %>%
arrange(geo_value, time_value) %>%
epiprocess::as_epi_df()
# Run model
arx_forecast <- lapply(
seq(7, 28, 7),
function(days_ahead) {
epipredict::arx_forecaster(
epi_data = target_data_arx,
outcome = "value",
trainer = linear_reg(),
predictors = "value",
args_list = arx_args_list(
lags = list(c(0, 7, 14)),
ahead = days_ahead,
quantile_levels = c(0.01, 0.025,
0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35,
0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7,
0.75, 0.8, 0.85, 0.9, 0.95,
0.975, 0.99),
nonneg = TRUE
)
)
}
)
# pull out the workflow and the predictions to be able to effectively use autoplot
arx_forecaster_workflow <- arx_forecast[[1]]$epi_workflow
arx_forecaster_results <- arx_forecast %>%
purrr::map(~ `$`(., "predictions")) %>%
list_rbind() %>%
mutate(horizon = forecast_horizon_wks,
location_id = state_name,
model = model_name,
target = target)
autoplot(
object = arx_forecaster_workflow,
predictions = arx_forecaster_results,
observed_response = target_data_arx %>%
filter(geo_value %in% geo_ids, time_value > (lubridate::as_date(forecast_date) - 4*30))) +
geom_vline(aes(xintercept = forecast_date))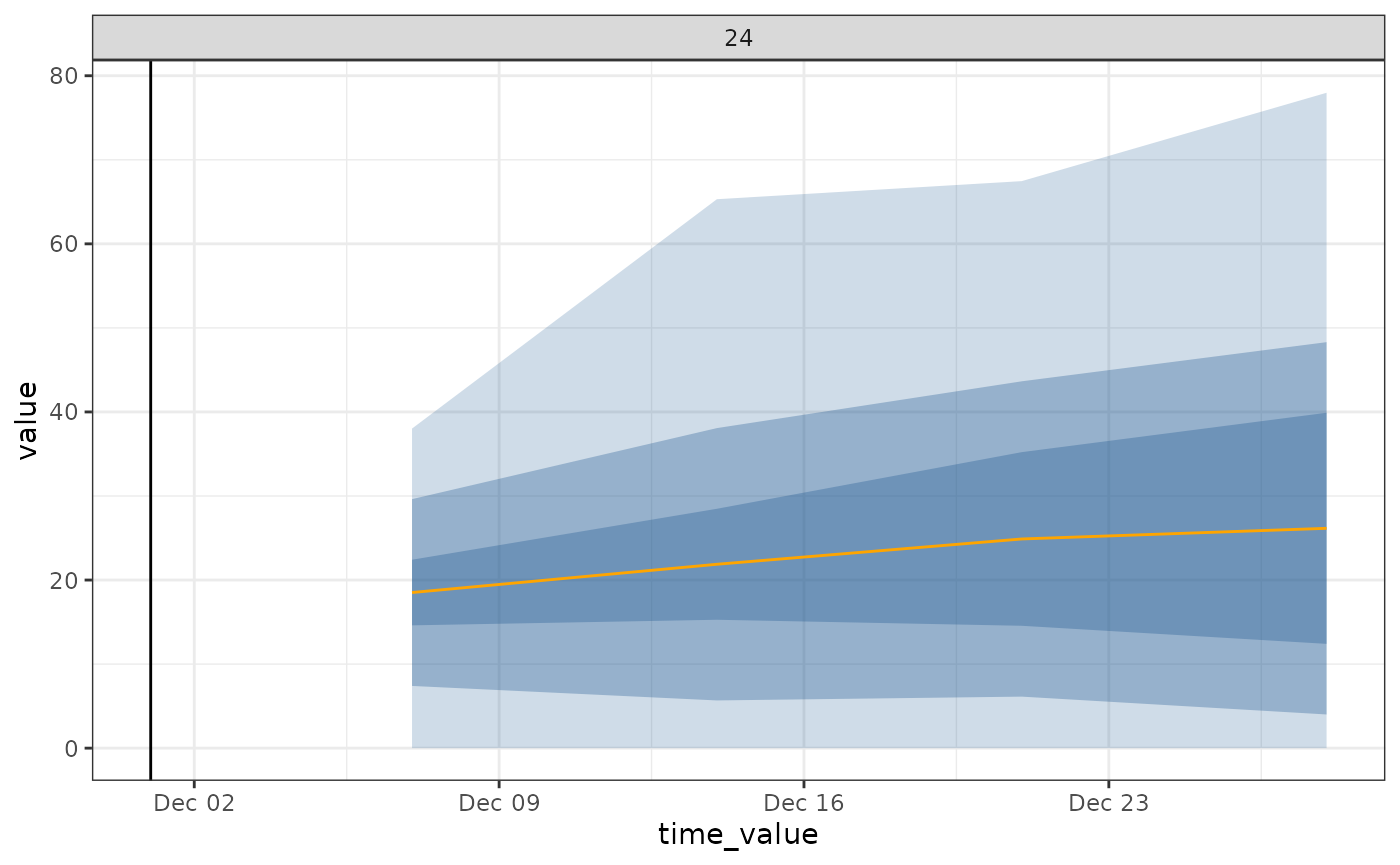
ARX model fit and forecast
Save in hubVerse Format
# Transform to hubVerse format
data_fc_arx_epipred <- trans_epipredarx_hv(fc_output = arx_forecast,
model_name = model_name,
target = target,
reference_date = reference_date,
horizon_time_steps = forecast_horizon_wks)
## Save data file
#' -- this will have validation build in for fluid workflow eventually.
save_model_output(model_name = model_name,
fc_output = data_fc_arx_epipred,
reference_date)## Model output saved to model-output/AMPH-epipredict-arx/2024-12-07-AMPH-epipredict-arx.csvClimatological Forecaster (epipredict package)
Here we use the epipredict package to fit a
climatological model to the data. We specify a forecast horizon of 7,
14, 21, and 28 days ahead (i.e., 1 to 4 weeks ahead).
Run the model
# Set up data for epipredict
target_data_clim <- target_data %>%
rename(geo_value = location,
time_value = target_end_date,
value = observation) %>%
tsibble::as_tsibble(index = time_value, key = c(geo_value)) %>%
arrange(geo_value, time_value) %>%
epiprocess::as_epi_df()
# Run the model
climate_forecast <- epipredict::climatological_forecaster(
target_data_clim,
outcome = "value",
args_list = climate_args_list(
forecast_horizon = forecast_horizon_wks,
time_type = "week",
quantile_levels = c(0.01, 0.025,
0.05, 0.1, 0.15, 0.2, 0.25, 0.3, 0.35,
0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7,
0.75, 0.8, 0.85, 0.9, 0.95,
0.975, 0.99),
center_method = "median",
quantile_by_key = "geo_value",
forecast_date = reference_date,
nonneg = TRUE
)
)
climate_forecast_workflow <- climate_forecast$epi_workflow
climate_forecast_results <- climate_forecast$predictions %>%
mutate(horizon = forecast_horizon_wks,
location_id = state_name,
model = model_name,
target = target)
autoplot(
object = climate_forecast_workflow,
predictions = climate_forecast_results,
observed_response = target_data_clim %>%
filter(geo_value %in% geo_ids, time_value > (lubridate::as_date(forecast_date) - 4*30))) +
geom_vline(aes(xintercept = forecast_date))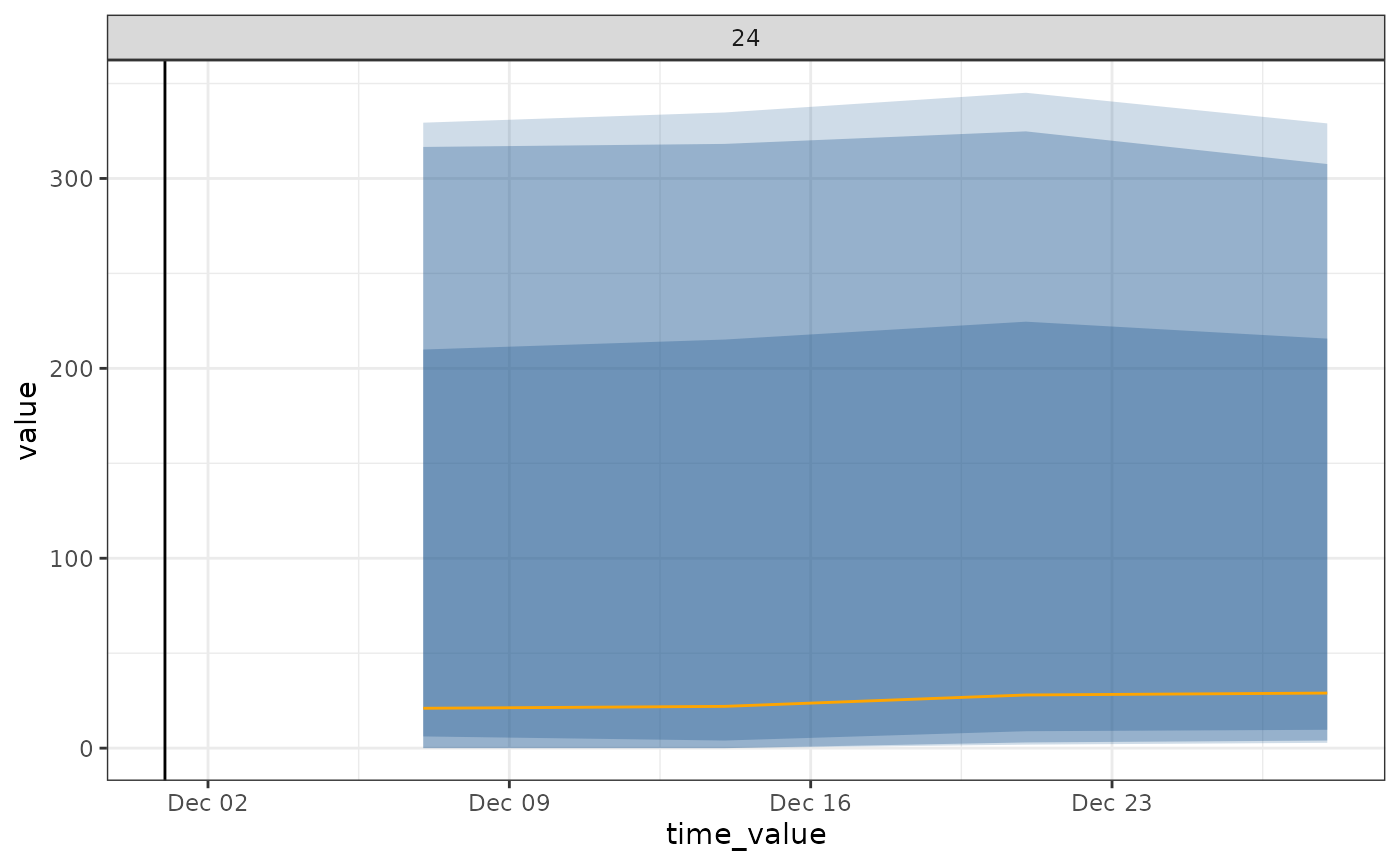
Climate model fit and forecast
Save in hubVerse Format
# Transform to hubVerse format
data_fc_climate_epipred <- trans_epipredclim_hv(fc_output = climate_forecast,
model_name = model_name,
target = target,
reference_date = reference_date,
horizon_time_steps = forecast_horizon_wks)
## Save data file
#' -- this will have validation build in for fluid workflow eventually.
save_model_output(model_name = model_name,
fc_output = data_fc_climate_epipred,
reference_date)## Model output saved to model-output/AMPH-epipredict-climate/2024-12-07-AMPH-epipredict-climate.csv